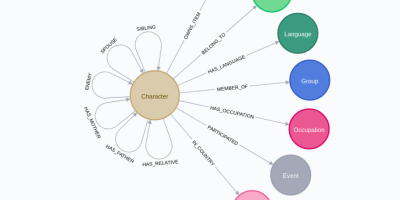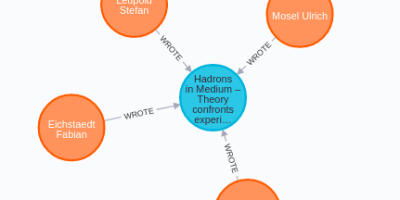
Centrality, Graph Algorithms, Graph data science, Neo4j, NLP

Centrality, Graph Algorithms, Graph data science, Neo4j, NLP
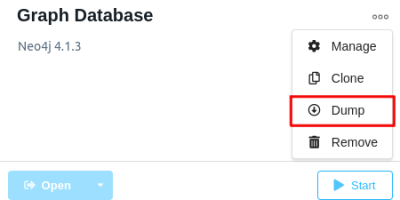
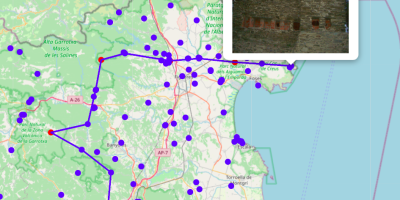
Graph Algorithms, Graph data science, mapbox, Neo4j, Spatial
Graph Algorithms, Graph data science, Neo4j, Spatial